Signed in as:
filler@godaddy.com
Signed in as:
filler@godaddy.com

Two open source Python library projects lead by Micron. They provide a new approach for building bespoke microscopes, in collaborations between biologists and physical scientists or engineers. Microscope and cockpit have been used to build several complex super-resolution microscopes as well as simpler microscopes.
Microscope-Cockpit (Cockpit) is a revolutionary new Python-based user friendly software interface for controlling and using bespoke microscopes built by physicists and engineers to tackle challenging biological questions. The code was based on the open source software developed to control the OMX microscope by John Sedat at UCSF Cockpit and is uniquely suitable for integration of advanced computational approaches including AI algorithms into the imaging process.
Python-Microscope (Microscope) is a revolutionary new Python library for controlling hardware devices, which can be used with cockpit, or independently of cockpit. Microscope consists of an abstracted set of drivers with a unified Pythonic grammar, to which devices according to specific “recipes”. Microscope hides the complexity of controlling some of the most advanced optical devices, such as adaptive optics and spatial light modulators as well as many cameras, high end piezo slip stick stages, lasers.

Three-dimensional structured illumination microscopy (3D-SIM) is a versatile and accessible method for super-resolution fluorescence imaging, but generating high-quality data is challenging, particularly for non-specialist users. We present SIMcheck, a suite of ImageJ plugins enabling users to identify and avoid common problems with 3D-SIM data, and assess resolution and data quality through objective control parameters. Additionally, SIMcheck provides advanced calibration tools and utilities for common image processing tasks. This open-source software is applicable to all commercial and custom platforms, and will promote routine application of super-resolution SIM imaging in cell biology. To download v1.0 click here
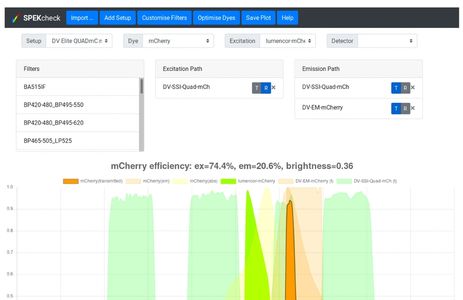
Advanced fluorescence imaging methods require careful matching of excitation sources, dichroics, emission filters, detectors, and dyes to operate at their best. This complex task is often left to guesswork, preventing optimal dye:filter combinations, particularly for multicolour applications. To overcome this challenge we developed SPEKcheck, a web application to visualise the efficiency of the light path in a fluorescence microscope. The software reports values for the excitation efficiency of a dye, the collection efficiency of the emitted fluorescence, and a "brightness" score, allowing easy comparison between different fluorescent labels. It also displays a spectral plot of various elements in the configuration, enabling users to readily spot potential problems such as low efficiency excitation, emission, or high bleedthrough. It serves as an aid to exploring the performance of different dyes and filter sets.
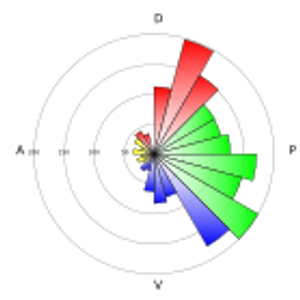
The study of dynamic cellular processes in living cells is central to biology and is particularly powerful when the motility characteristics of individual objects within cells can be determined and analysed statistically. However, commercial programs only offer a very limited range of inflexible analysis modules and there are currently no open source programs for extensive analysis of particle motility. Here, we describe ParticleStats (www.ParticleStats.com), a web server and open source programs, which input the X,Y co-ordinate positions of objects in time, and outputs novel analyses, graphical plots and statistics for motile objects.

The primary source of funding for our cutting-edge imaging research.
Principal Applicant: Ilan Davis
Co-applicants: Jordan Raff, Martin Booth, Yvonne Jones, Christian Eggeling, David Stuart, Kay Grunewald, Neil Brockdorff.

Funded by the Wellcome Institutional Strategic Support Fund and the John Fell Fund, these two innovative microscopy development projects, the 4Pi SMS & Microscopi, are aimed at very different areas of research.

Nanoscopy Oxford (NanO), a new initiative funded primarily by the MRC to develop novel super-resolution imaging modalities to address key questions in the field of biomedical research.